Whole cell research
The functions of lives are kept by hierarchical structures of molecules that are organizing lives themselves, so that it is essential for understanding lives to analyze the spices and the number of synthesized molecules and the time-series of molecular synthesis quantitatively. Previously, spatial and structural studies of proteins in living cells were performed by using solution NMR and fluorescent microscopy; however, it was difficult to measure and quantify all the molecules in living cells.
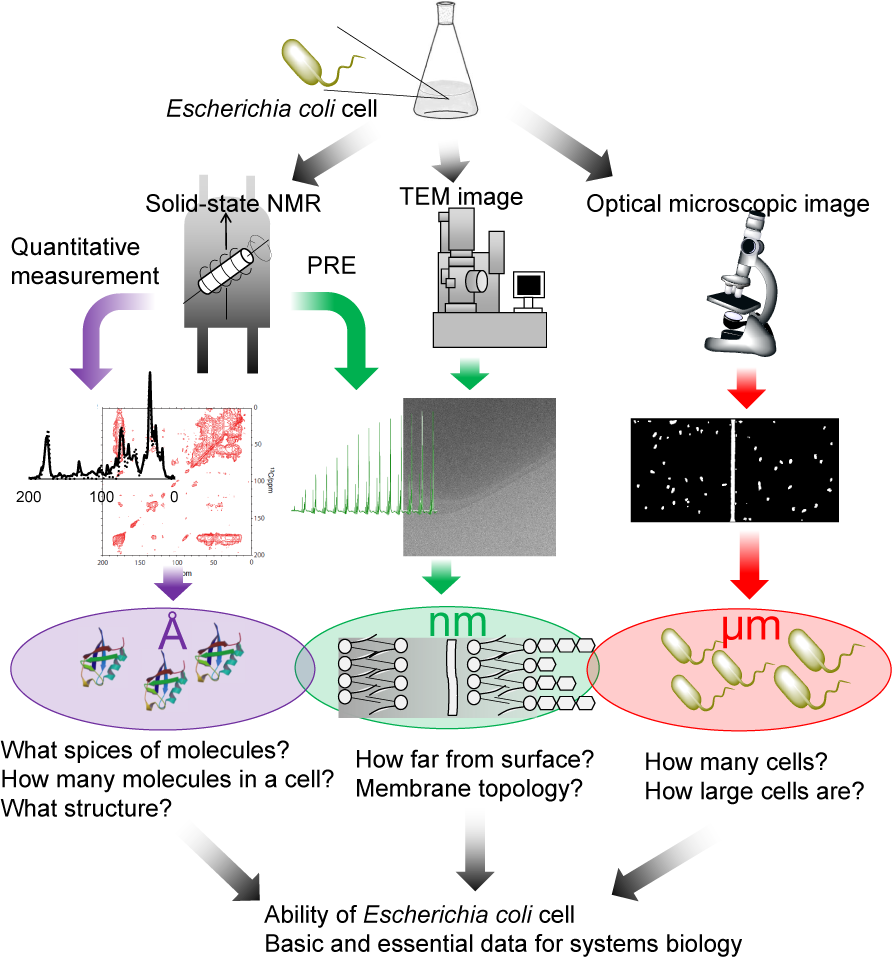
We focused on solid-state NMR to obtain such quantitative data. The first character of solid-state NMR is the ability that can observe all the molecules in cells including huge complexes like membrane proteins especially under low temperature. Secondly, it does not have any invasiveness. That means it can detect molecules inside cells without breaking any cells or proteins so that we can obtain information inside cells that are still reflected from their living phenomena. Finally, NMR has the quantitative behavior in principle so is used as qNMR for measuring the purity of chemical compounds, for instance. That means the signal intensities are completely proportional to the number of molecules. We aimed to quantify the molecules inside a cell and approach the function of living organisms by combining such characteristics of solid-state NMR. Moreover, we also combine the atomic resolved information obtained from solid-state NMR and higher ordered structural information of a cell obtained from optical and electron microscopy, then close in a cell itself consisted of multiscale structures.
What kind of life phenomena can we reveal by using cellular solid-state NMR? Honestly, it is difficult for new methods to apply for complex human or mammalian cells. Thus, we focused on Escherichia coli (E. coli) cells as a model organisms and ubiquitin overexpression as a model phenomenon for the validation of new methods. At first, we compered the experimental NMR spectra of E. coli cells and simulated NMR spectra of known molecular components of E. coli cells, and then clarified the molecular spices synthesized during the ubiquitin overexpression. Next, we succeeded to count each molecule by counting spins from spectral intensities.
Not only the number of molecules but also structural and spatial information of molecules in cells was obtained from cellular solid-state NMR measurements. We would like to introduce one example of structural study of proteins in E. coli cells. In the process of molecular counting research, we succeeded to separate the signals originated from ubiquitin in E. coli cells. The comparison of the chemical shifts between the spectra of ubiquitin in E. coli cells and the simulated spectra of ubiquitin proved that the structure of ubiquitin in cells still form the same structure of ubiquitin in vitro. In another example is obtaining spatial information of molecules in cells. We succeeded to discriminate between cytoplasm and extracellular space by using paramagnetic molecules. In future, we will able to know where the synthesized molecules are in a cell.
One of the weak point of NMR is to obtain just the average information of the whole sample. In order to see the distributions of E. coli cells, we calculated the size-distribution of cells and counting the number of cells in the media from the optical microscopic images. Moreover, we showed the changes of the size and the number of the cells were consistent with the developments of the number of molecules obtained from solid-state NMR measurements. In this manner, we are approaching the nature of E. coli cells by combination of solid-state NMR and microscopes.
You may think what the significance of obtaining such data is. For instance, you can calculate the volume ratio of synthesized ubiquitin in a cell and the yield of ubiquitin in 1 L culture media. Moreover, you may be interested in how many ubiquitin molecules a ribosome synthesizes per one second. You can also know the incorporating ratio of glucose, and how much percentage such glucose is involving into ubiquitin or discharging as carbon dioxide. This knowledge obtained from the research should be essential for estimating the ability of E. coli cells and useful for promoting systems biology of E. coli cells.