
Japanese
Laboratory of
Structural Proteomics
Institute for
Protein Research
Osaka
University
3-2 Yamadaoka,
Suita, Osaka 565-0871, JAPAN
FAX (from
abroad) +81-6-6879-8599
Associate
Professor : Takahisa IKEGAMI, Ph.D.
E-mail
: tiik@protein.osaka-u.ac.jp
The
group is studying biomolecules such as proteins, nucleic acids, and lipids
mainly by two methods, X-ray crystallography and nuclear magnetic resonance
(NMR).
1. Nuclear
Magnetic Resonance
NMR is a useful tool for the analyses of protein
structures and dynamics with atomic resolutions. This group determines the three-dimensional structures of proteins using
NMR, studies dynamics of proteins, and develops methodology of related NMR
techniques.
2. Determination of Protein Structures
It has been more and more important to describe biological
functions in terms of the three-dimensional structures of associated proteins.
At least three methods are known to determine structures with the atomic coordinates:
X-ray crystallography, electron-microscopy, and NMR. NMR is characteristic in a
point that it can analyze the structures of biomolecules in solution states,
providing data that reflect more natural conditions. Therefore, NMR allows for
the analyses of proteins containing a lot of flexible regions, for which
crystallizations are generally difficult. Although it is considered that the
procedures of determining the structures of small proteins (M.W. < 30 kDa)
by NMR have mostly become protocols, a lot of problems have to be practically
solved when flexible and large proteins (M.W. > 40 kDa), such as ones
containing multi-domains inside, are targeted.
3. Analyses of Protein Dynamics
NMR is
also suitable for detection of flexibility, providing information as to which
parts of proteins fluctuate in solutions. The fact that NMR yields dynamic
structures can distinguish NMR from X-ray crystallography, which normally gives
static structures with higher resolutions. A lot of biological functions are
related to protein-motions. In particular, slow dynamics occurring in a time
range of microsecond to millisecond are very important to biological functions.
Since much faster motions ranging from picosecond to nanosecond have been
mainly analyzed by NMR so far, new parameters representing slow dynamics are
expected to elucidate new characters of proteins as well as mechanisms of
protein folding/unfolding and protein-protein associations.
4. Development of NMR Methodology
Further developments of NMR methodology to facilitate the
above-mentioned studies are also important tasks. For example, new methods for
structure determination of much larger proteins with molecular weights of >
40 kDa and for observation of slow dynamics by NMR should be developed more.
 |
|
Fig. 1 The super-conducting-magnet
in a 600 MHz nuclear magnetic resonance (NMR). A cooling medium, liquid helium,
is just being transferred to the magnet to keep the temperature of the
super-conducting coils at 4 K. The magnet functions like a Dewar thermos
vessel, in which a phase of liquid helium containing the coils is surrounded by
that of liquid nitrogen. |
 |
Fig. 2 The top part of an 800 MHz
NMR magnet. A solution sample stuffed at the bottom of a glass tube is just being
placed in the magnet. |
|
 |
Fig. 3 The spectrometer part of an
NMR. It generates radio-frequency waves in forms of pulses, emits them to the
sample in the magnet, processes the signals having returned back from the
sample, and saves these data. |
|
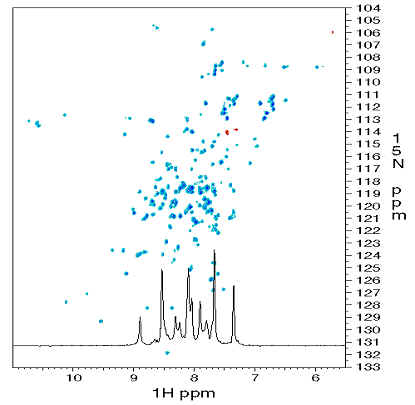 |
Fig. 4 A two-dimensional NMR
spectrum of a protein. The spectrum is referred to as 1H-15N
heteronuclear single quantum correlation (HSQC), in which each peak corresponds
to the 1H and 15N resonances of the amide group in an
amino acid. These kinds of spectra imply the stabilities in conformations of
proteins. |
The 1H-15N pulse sequences (Bruker) for
the participants in JASS'03 NMR-Winter-School (2004/Jan./22nd).