|
revised February 09, 2022

|
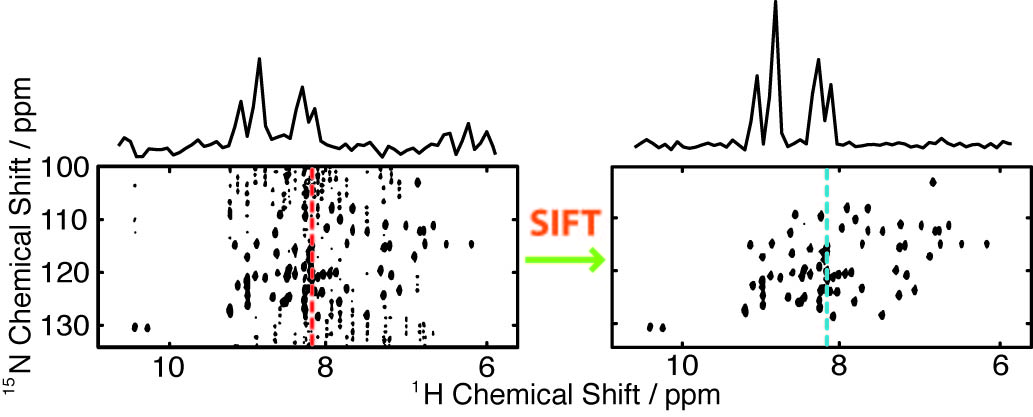
The SIFT process interleaves fast Fourier transforms back and forth between the time and frequency domains with applications of experimental data in the time domain and known zeroes or ''dark points'' in the frequency domain. In this manner, SIFT can fill in the time data points that are missing due to non-uniform sampling (NUS) or removed due to experimental problems.
In the sampling-limited cases, NUS enabled by SIFT can expedite high resolution multidimensional acquisition, by reducing the number of samples required at long evolution times.
In the sensitivity-limited cases, NUS enabled by SIFT can improve signal-to-noise by sampling more heavily early in each FID.
2-D, 3-D time domain datasets completed by SIFT may be output in nmrPipe format and Sparky format for further manipulation.
| SIFT macros | version |
|---|---|
| Suite of MATLAB macros for 2D SIFT | 2.4 |
| Suite of MATLAB macros for 3D SIFT | 2.4 |
| SIFT manual | Flow chart |
|---|---|
| 2-D SIFT Stepwise manual (PDF) | flow chart (PDF) |
| 3-D SIFT Stepwise manual (PDF) | flow chart (PDF) |
When you use SIFT, please cite:
Yoh Matsuki, Matthew T. Eddy, and Judith Herzfeld (2009).
Spectroscopy by Integration of Frequency and Time domain information (SIFT) for Fast Acquisition of High Resolution Dark Spectra.
J. Am. Chem. Soc. 131, 4648-4656.
For an example of an application to solids, please see:
Yoh Matsuki, Matthew T. Eddy, Robert G. Griffin, and Judith Herzfeld (2010).
Rapid Three-Dimensional MAS NMR Spectroscopy at Critical Sensitivity.
Angew. Chem. Int. Ed. 49, 9215-9218.
For an example of an application to serial relaxation measurements, please see:
Yoh Matsuki, Tsuyoshi Konuma, Toshimichi Fujiwara, and Kenji Sugase (2011).
Boosting Protein Dynamics Studies using Quantitative Nonuniform Sampling NMR Spectroscopy.
J. Chem. Phys. B 115, 13740-13745.
Start up MATLAB, edit the argument section in the SIFT run macro (either sift2d_v2_4.m or sift3d_v2_4.m), and implement commands sequentially. All specific instructions are given in the macros.
The suite of MATLAB macros for 2-D and 3-D SIFT processing were coded and tested in MATLAB version R2012b. They are available for downloading at
http://www.protein.osaka-u.ac.jp/matsuki/SIFT.
The SIFT-processed data can be output in nmrPipe format (http://spin.niddk.nih.gov/NMRPipe/)
and Sparky format (http://www.cgl.ucsf.edu/home/sparky/)
for inspection, analysis and comparison of spectra.
The only requirement for SIFT sampling is that the points be chosen from a uniform grid to accommodate fast Fourier transforms.
It is known that the amplitude of the pseudo-noise from NUS is proportional to the coherence in the sampling pattern (J. C. Hoch et al., JMR 193, 317-320, 2008).
However, generating incoherent schedules via schemes that start from randomly chosen seeds yields irreproducible results of variable quality.
A simple, general, and easily reproducible method for generating non-uniform sampling schedules that preserves the benefits of random sampling is described by
Matthew T. Eddy, David Ruben, Robert G. Griffin, and Judith Herzfeld (2012).
Deterministic schedules for robust and reproducible non-uniform sampling in multidimensional NMR.
J. Magn. Reson. 214, 296-301, 2012.
An ASCII text file of the time table used for SIFT processing should contain a series of integers in one (for 2D) or two (for 3D) columns that corresponds to multiples of a fixed dwell time for each evolution dimension. The sequential order of the schedule should be matched to the order of stored FIDs in the dataset to be processed.
This software is provided "as is", and you cannot re-distribute it. The authors make no warranties, either express or implied, as to any matter whatsoever with respect to the software. In no event will the authors be liable for any loss of profits, or any incidental, special, exemplary, or consequential damages of any nature whatsoever, arising out of or relating to the use or performance of the software.
If you have any comments, suggestions, concerns or questions, please contact us.