1) SAIL-NMR法を活用した新しい蛋白質構造動態研究法 の開発
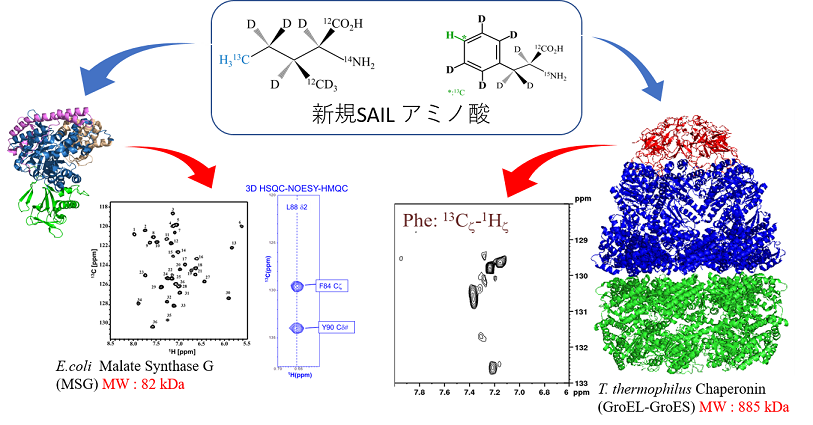
NMR法の最大の問題点である “分子量の壁”、“低感度” を克服するため、SAILアミノ酸の改良を進めている。現在、82 kDaのリンゴ酸合成酵素(MSG)や468 kDa のアミノペプチダーゼ複合体 (TET2) 及び 1000 kDaに及ぶGroEL-GroES複合体を対象に各種SAILアミノ酸を導入し、NMRシグナルの高感度検出、シグナル帰属法および溶液構造解析法の開発を進めている。
Structural studies of larger proteins using new SAIL amino acids
At present, the solution NMR spectroscopy of large molecular proteins (>50 kDa) relies exclusively upon the information obtained from the backbone NH and side-chain CH3 signals, which are not sufficient for precise structural studies. In order to overcome this situation, we have been trying to further optimizations of the isotope labeling patterns for stereo-array isotope labeled (SAIL) amino acids. We introduce a precise structural analysis of 80 – 900 kDa large molecular protein and protein-protein complexes (Malate Synthase G; TET2 Aminopeptidase, and GroEL-GroES complexes) by using the relaxation optimized SAIL-NMR method.
関連論文:
Miyanoiri et al., BBA. Gen. Subj. (2020)
[https://doi.org/10.1016/j.bbagen.2019.129439]
Gauto, … Miyanoiri et al., JACS (2019)
[https://doi.org/10.1021/jacs.9b04219]
Kainosho, Miyanoiri et al., J. Biomol. NMR (2018)
[https://doi.org/10.1007/s10858-018-0198-x]