RESEARCH
Three-dimensional protein structure
brings us a deeper insight into the biological function. X-ray crystallography is the best method to determine atomic coordinates of protein molecules. The main aim of our group is the X-ray structure determination of the biological macromolecular assemblies including membrane protein complexes in order to elucidate the molecular mechanism of the highly organized biological processes at atomic level.
1. Structural studies of photosynthetic membrane protein complex and related redox enzymes.
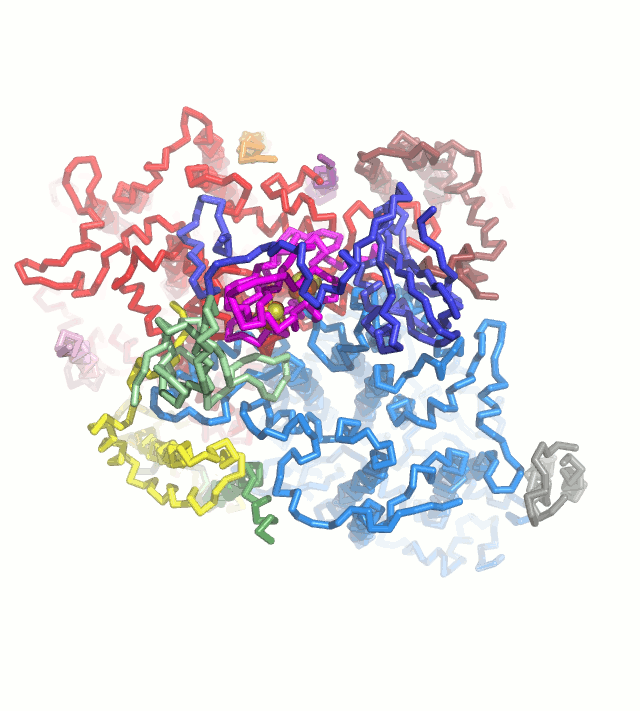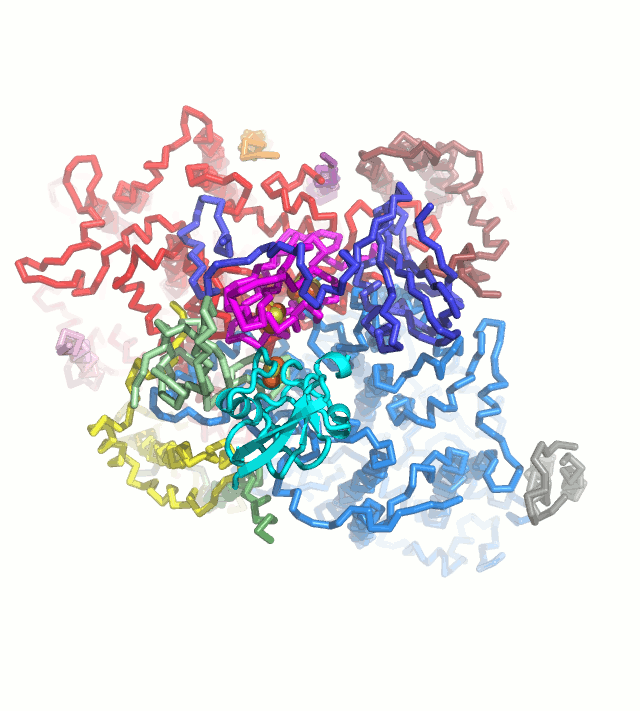
Photosynthetic light reaction establish electron flow in the chloroplast thylakoid membranes, leading to the production of ATP and NADPH, and also providing the reducing power to many redox enzymes. We want to understand the blanching mechanism of this electron flow based on the crystal structures. Ferredoxin (Fd) is a key protein that is reduced from photosystem I and serve as an electron carrier protein for many Fd-dependent enzymes, including Ferredoxin-NADP+ reductase, Sulfite reductase, Nitrite reductase, Glutamate synthase (Fd-GOGAT), Protochlorophyllide reductase and Hydrogenase. These Fd-dependent enzymes do not have any consensus sequence and common structural motif. We are trying to crystallize these Fd-dependent enzymes complexed with Fd in order to provide the structural basis for Fd-dependency .
2. Crystal structure analyses of dynein motor
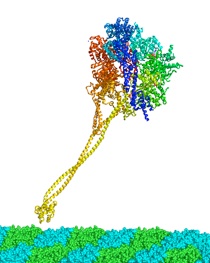
Dynein is a microtuble-based motor protein, consisting of the identical heavy-chains with assorted light-, light intermediate- and intermediate chains. The motor activity is located in the heavy chain, whose molecular mass is more than 500kDa. Sequence analysis and electron microscopy reconstuctions indicate that the microtuble-binding domain of dynein heavy chain is separated from the AAA core of the motor which contains the ATP hydrolysis sites, by an elongated stalk domain consisting of an anti-parallel coiled-coil structure. It was hypothesized that the dynein utilized small amounts of sliding displacement between the AAA core and the microtuble-binding head. However, the structural basis of how to slide the two long colied-coil helices in the opposite directions and couple the microtuble binding is still unkown. In order to address these questions, we are trying to crystallize the several recombinant proteins of the dynein stalk.
3. High resolution and damage-free structure analysis of metalloproteins
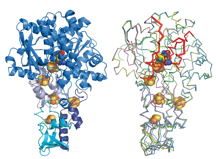
Characteristic property of plant-type Fd is a very negative redox potential (-420 mV for maize leaf Fd), therefore it is purified as an oxidized form through normal purification procedure. Although it can be chemically reduced, solution or crystal sample is immediately oxidized by molecular oxigen. Most of the reported X-ray structure are believed to be in oxidized form. However, we would like to pointed out that, when using the frozen crystals of Fd for X-ray data collection, the dark brown color of the frozen crystal is obviously changed to light brown during X-ray beam irradiation at Synchrotron. Also, honestly speaking, noisy peaks of difference fourier map around the [2Fe2S] cluster is often very hard to disapper after enough cycles of crystallographic refinement of plant-type Fd. It may suggest that currently reported structure was a time-averaged structure of partially reduced (light brown) and oxidized (dark brown) Fd, which is crystallographically very difficult to be resolved. In order to elucidate the radiation-free structure of plant-type Fd crucial to understand the structural change of Fd upon reduction, further diffraction study of the plant-type Fd is necessarily needed.

