Introduction
Cells, the smallest unit of living organisms, have a sophisticated mechanism for determining their fate. This mechanism can be imagined as a network resembles an electric circuit that connects genes and proteins. However, even after the entire human genome has been decoded, it is still difficult to identify all connections in networks containing more than 20,000 genes. As one of the showcases of these networks, we study signal transduction networks and clarify the relationship between the network structure, that is regulatory rules, the dynamics of molecules in the network and the cell fate.
Research interests
Our Laboratory has been carrying out researches to identify the regulatory principles of cell fate decisions by computational, theoretical and experimental approaches. For this purpose, we employ both bottom-up mechanistic mathematical modeling and top-down omics methods. These methods complement each other to understand the cells at the system level.
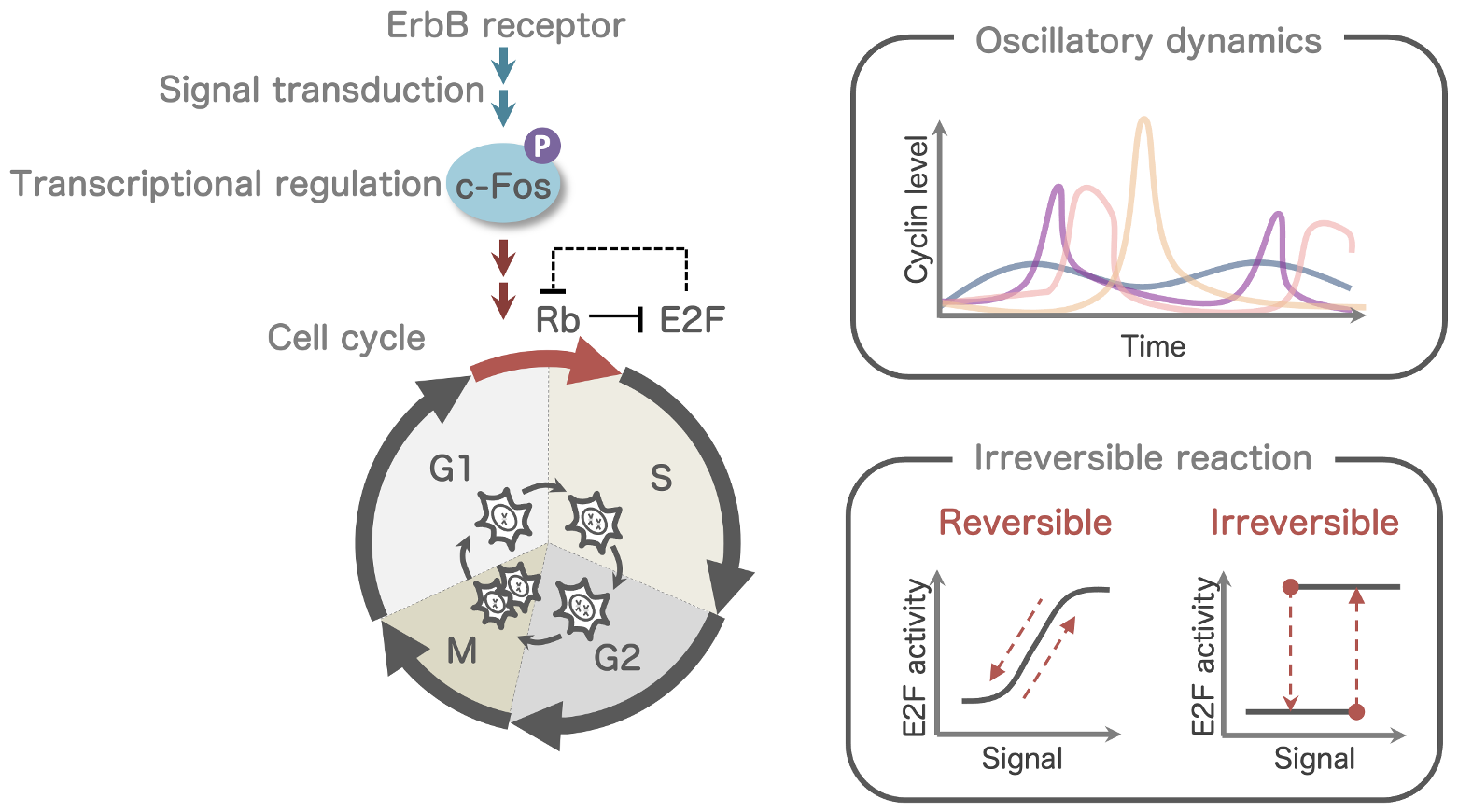
ErbB signaling network and cell fate regulation
- Cell cycle regulation
- Early responsive gene regulation
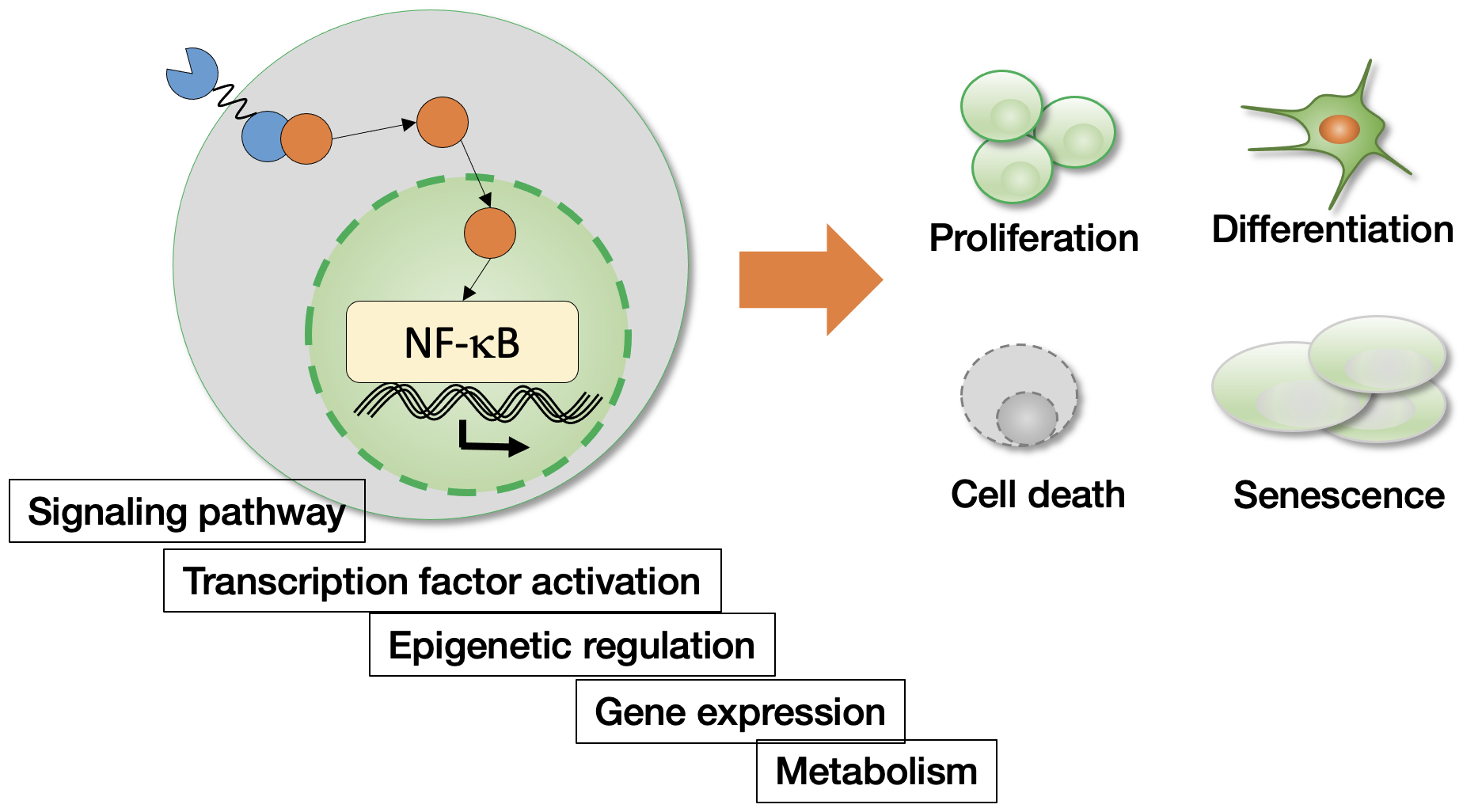
NF-kB signaling network and cell fate regulation
- Immune cell regulation
- Aging
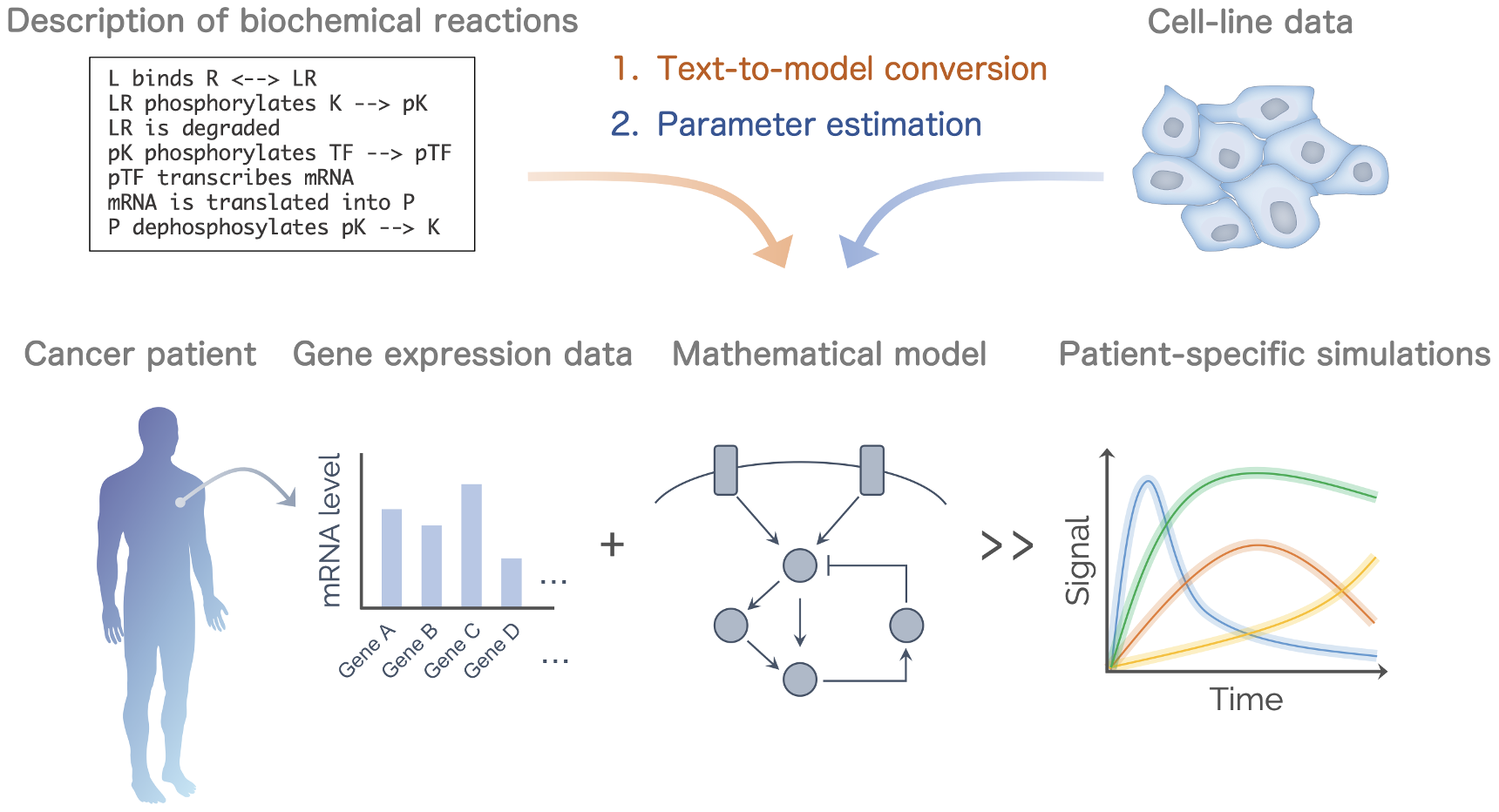
Development of modeling and informatics framework
- Patient-specific model for stratification of cancer
- Natural language processing (NLP) and text-to-model for mathematical knowledge-free modeling
- Bioinformatics pipelines for single cell transcriptome and epigenetics