We are looking for students
The laboratory for Cell Systems is hiring students who are interested in biology and/or programming.
Students from any background (Biological sciences/Information sciences/Engineering) are welcome.
The research topics in our lab are as follows:
1. |
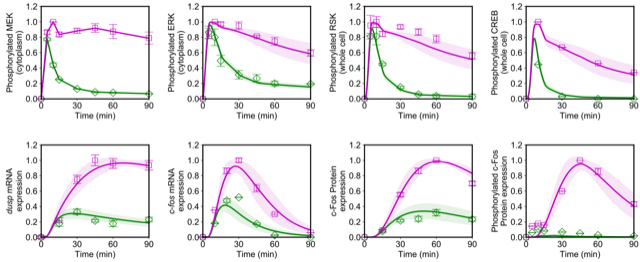
Modeling of Biological Systems → Okada lab mathematical models
Mathematical model to infer underlying mechanisms of dynamic cellular control from experimental data.
|
|
2. |
Imaging based cellular dynamics analysisConfocal and widefield fluorescence microscopy based imaging to observe transcription factor dynamics and cell cycle progression. Automation of image processing and analysis.
|
|
3. |
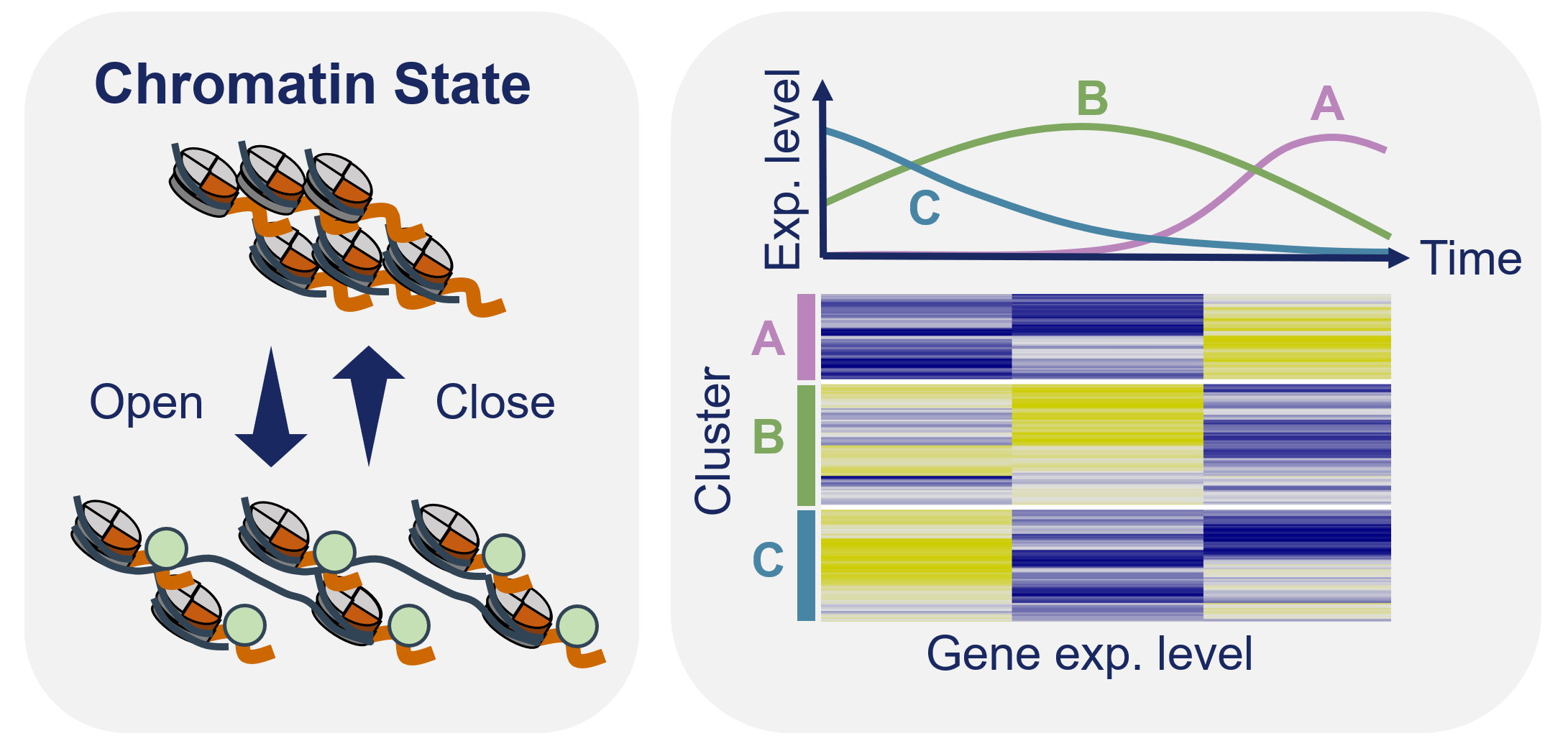
Omics analysisComprehensive data analysis of whole-genome gene expression, histone modification and chromatin structure to clarify new gene expression regulation systems.
|
|
4. |
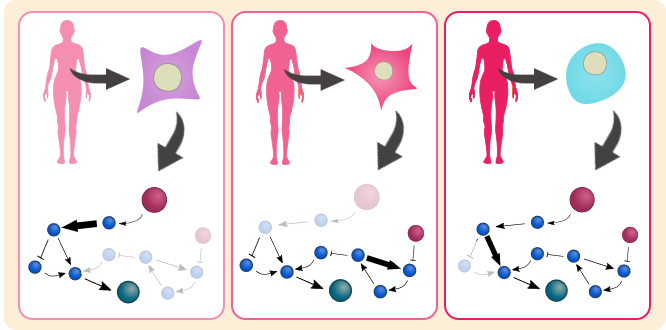
Mathematical analysis of clinical dataClarify the mechanism of signaling network using TCGA database and develop a new method to classify the cancer subtypes.
|
|